Our Research
Why HLA-DQ?
Part 2: Understanding HLA-DQ Antibodies
Antibodies can recognize epitopes on one or both chains of HLA-DQ
Prior to molecular methods, patients were typed for HLA-DQ (and -DR) using antisera reagents that differentiated DQ molecules into groups that shared similar ꞵ-chain subunits, termed serotypes. It is now considered to be a low-resolution typing method, as even today’s serologic reagents are only capable of discriminating between seven HLA-DQ serotypes, still identified by the molecule’s ꞵ-chain: DQ2, and DQ4-9. Despite the development of molecular-based methods that allowed for intermediate- and high-resolution typing of the DQA and DQB loci, HLA-DQ antigens (and antibodies) continued to still be routinely reported using serologic nomenclature. But because HLA-DQ serologic naming convention only communicates the molecule’s ꞵ-chain typing with disregard to its α-chain, this falsely gave the impression that patients develop antibodies mostly against the ꞵ-chain of DQ molecules.
HLA nomenclature can be confusing! So confusing that even the nomenclature to describe the nomenclature can be confusing too. This figure lays out the different levels of HLA-typing, ordered from highest (top) to lowest (bottom) resolution. While the bold terms are usually the most frequently used phrases, you may sometimes hear different vocabulary to describe the same thing. Some of these terms are included within their appropriate level of the pyramid.
CASE STUDY
Our suspicions that DQα’s involvement in antibody formation can be clinically profound began with a case involving an unexpected negative crossmatch in June of 2006. With the rise of the virtual crossmatch beginning around this time, patient John Doe was assessed for vXM using the following typing information:
Patient Typing: A2 A26 B44 B60 DR4 – DQ8
Donor Typing: A2 A3 B7 B60 DR4 – DQ7 DQ8
Importantly, although the SAB assay was being introduced around that time, our laboratory was still using the Single Antigen Flow-PRA assay, which had only 7 beads coated with HLA-DQ antigens: one bead for each serologic specificity. Results of Class II antibody specificities by Flow PRA beads revealed strong donor specific antibodies (DSA) against DQ7.
Based on these data, it was determined that the DQ7 DSA would likely yield a positive physical crossmatch for this donor. However, upon completion of a flow cytometry crossmatch, it was found that both B- and T- cell FCXM results were negative. In order to investigate this discrepancy, we decided to run the SAB assay (which was just being introduced into our clinical lab), perform high-resolution typing on the donor DQA/DQB, and obtain the typing information for the DQ7 bead used in the Flow-PRA Single Antigen assay from the vendor. The results of these efforts are as follows:
While the patient exhibited antibodies against four out of the five DQ7 specificities on the SAB panel, he did not show reactivity with the DQ7 antigen containing a DQA*03:01 α-chain. High-resolution typing of the donor and Flow PRA bead revealed a DQ7 phenotype of DQA1*03:01~DQB1*03:01 and DQA1*05~DQB1*03:01, respectively, explaining the initially discrepant positive Flow PRA antibody screen and negative FCXM.
It becomes clear that using DQ serologic nomenclature is not sufficient when reviewing antibody and typing information for virtual crossmatch assessments. In the discussed case, it was assumed that the patient possessed DSA against the potential donor when in fact he did not. If this phenomenon – that patients could develop antibodies directed against only a subset of a serotype defined by specific α-chains – proved to be more prevalent, serologic nomenclature would be unable to capture this. The next question, naturally, then became “how common is this?”
Antibodies don’t care what serotype you are, but they do care about which heterodimers you have
In the late 2000’s, we reported numerous cases where patients exhibited anti-DQ antibodies that recognized epitopes located on the α-subunit of the molecule, as well as epitopes that are likely generated by the combination of both alpha and beta chains. When using serologic nomenclature, this can sometimes appear as though a patient possesses antibodies against their own DQ molecules. However, closer inspection of the beads would reveal that the “self” reactivity is confined only to a subset of the patient’s serotype – those coated with the patient’s own DQꞵ typing but paired with non-self α-chain(s). This phenomenon was observed so frequently, in fact, that in 2010 we reported that out of the 104 kidney waitlist patients at our center with known anti-DQ antibodies, nearly three-quarters (71%) of them possessed antibodies reactive against SAB coated with either the α- or ꞵ- chain of their own DQ typing.
Figure: Examples of SAB assay results for patients with DQ antibodies directed against their own ꞵ-chain (left) or their own α-chain (right). The patient’s typing is listed at the top of each table, with bead reactivity information shown underneath. A bar graph showing MFI values for each bead is shown to the right of each table. Beads that had MFI values over 1000 are highlighted in yellow. Beads with the patient’s own DQB typing that showed positive reactivity are highlighted in blue, and beads with the patient’s own DQA typing that showed positive reactivity are highlighted in red. 34% of patients in our study exhibited antibodies against beads coated with their own DQꞵ chain when combined with a non-self α-chain. Likewise, 62% of patients exhibited antibodies against beads coated with their own DQα chain when combined with a non-self ꞵ-chain.
Adapted from: Tambur AR, Leventhal JR, Friedewald, & Ramon DS. 2010. The complexity of human leukocyte antigen (HLA) DQ antibodies and its effect on virtual crossmatching. Transplantation 90:1117-24
The serologic nomenclature of Class II HLA molecules is derived from the typing of the ꞵ-chain. Consequently, molecules with non-self-DQα paired with self-DQꞵ have the same serological typing as self-DQα paired with self-DQꞵ. We were able to show in this study that patients are capable of developing antibodies with specificities directed toward DQ molecules that share the patient’s own DQꞵ typing when combined with a non-self α-chain. Yet, for many years it was common practice to report antibody specificities using serologic nomenclature. Even today, UNOS only allows DQB1 assignment at the serologic level and disregards DQA1 for cPRA and allocation point calculations. The ramifications of this can be profound. For example, waitlist patients that possess DQ antibodies to only a subset of a particular serologic group could be denied potential compatible donors because the entire DQ serotype was blocked in UNOS. [Read more about OPTN allocation policy here]
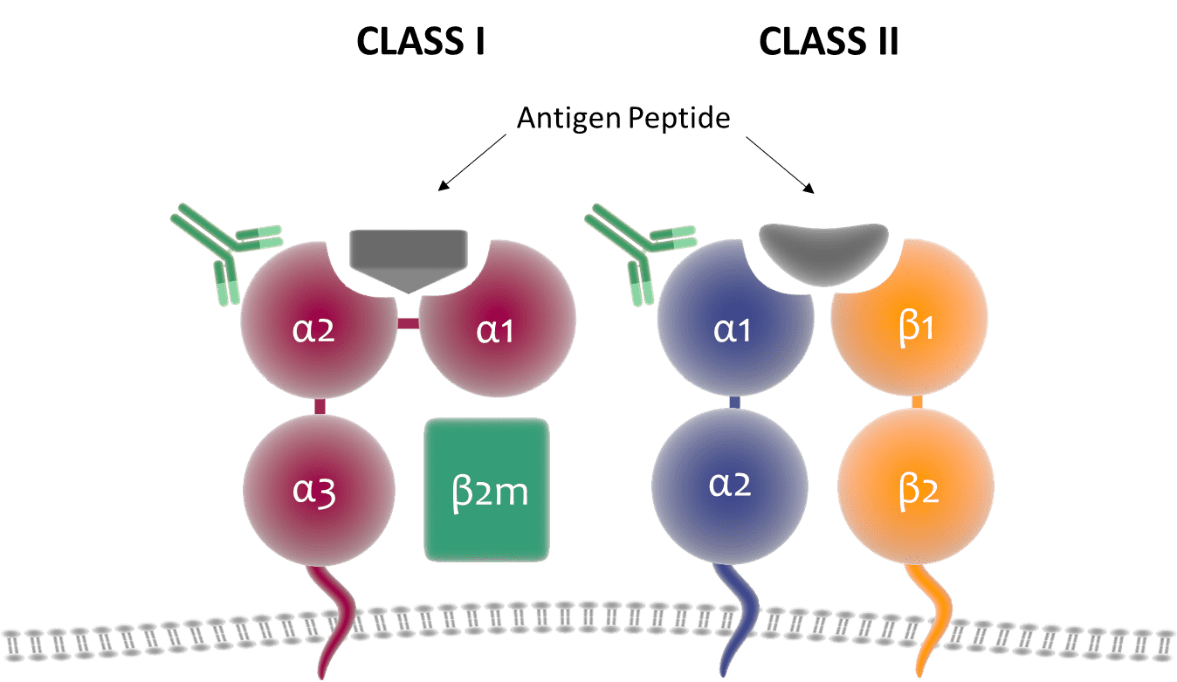
We were also able to show that antibodies can target DQ molecules that share the patient’s own α-chain if paired with a non-self-ꞵ-chain. Although it does not lead to the same antibody assignment challenges as discussed in the preceding paragraph, it does encourage further exploration of DQ molecular structure and how it is targeted by antibodies. Here we’ll need to revisit the genetics and structure of HLA molecules. Class I heterodimers are encoded by a polymorphic α-chain (which itself is composed of the α1, α2, and α3 domains) and ꞵ2-microglobulin. Class II heterodimers, on the other hand, are encoded by an α- and ꞵ-chain, both of which are polymorphic, apart from DRA1. In Class I molecules, the α1 and α2 domains comprise the extracellular region of the protein and have been shown to be targeted by anti-HLA antibodies This region is homologous to the outer domains of the α- and ꞵ-chains of Class II molecules, yet serology naming convention is dictated only by the ꞵ-chain. While a non-issue for HLA-DR, it would be reasonable to assume that the polymorphic α-chains of HLA-DQ and -DP contribute to the overall immunogenicity of those antigens and can thus induce antibody production.
Figure: Structure of HLA Class I and Class II molecules on the cell surface. CI antigens are comprised of a polymorphic α-chain and conserved ꞵ-2-microglobulin protein, while CII antigens include a polymorphic α- and ꞵ-chain. CI α-chains contain three domains – the α1 and α2 domains make up the outer most extracellular region and is homologous to the α1 and ꞵ1 domains of CII α- and ꞵ-chains.
If the CII α-chain can drive antibody production, how can we indicate this with serologic nomenclature? (Answer: we can’t!)
From these observations, it becomes evident that unlike HLA-DR antigens, which enjoy a monomorphic α-chain and therefore do not fall victim to the same serology-based typing obstacles, testing for DQ antibodies must consider both α- and ꞵ-chains of the molecule. Furthermore, it is important to keep in mind that the DQ molecule expressed on the cell surface is a heterodimer, a single molecule comprised of an α- and ꞵ-chain, and that the epitopes recognized by antibodies are likely influenced by the three-dimensional structure created by both subunits.
Antibody recognition sites are likely a product of both α– and ꞵ– chains
In the early 2000’s Rene Duquesnoy developed HLAMatchmaker, a program that computed the number of polymorphic differences between HLA mismatched pairs in the form of eplets, small clusters of amino acid residues within a radius of about 3 Ångstroms. The software was developed based on the working theory that the functional epitopes of HLA molecules can be characterized by these eplets, and that the degree of eplet incompatibility between a recipient and donor influences how immunogenic the donor graft will be. Using this software in conjunction with Cn3D from NCBI, we showed in 2014 that although eplets may be located on either the α- or ꞵ- chain of DQ molecules, the antibody “footprint”, the 15 Ångstrom area representing the full structural epitope recognized by an antibody, more likely than not covers an area encoded by both chains. Taken with MFI information from SAB assays, we additionally showed that patients who had multiple antibody specificities sharing a common eplet displayed a broad range of MFI values, indicating a heterogeneity in antibody binding affinity. We proposed that this could be explained by subtle differences in the three-dimensional conformation caused by variations in DQα/DQꞵ paring.