 Triple Z joins our lab for his Master of Science in Biotechnology. Look at all the sunshine he brings!
Triple Z joins our lab for his Master of Science in Biotechnology. Look at all the sunshine he brings!
 Our research on chemical mapping in mouse ES cells is highlighted in Nature Reviews Molecular Cell Biology. Read it here: The chemical brothers: nucleosomes and transcription!
Our research on chemical mapping in mouse ES cells is highlighted in Nature Reviews Molecular Cell Biology. Read it here: The chemical brothers: nucleosomes and transcription!
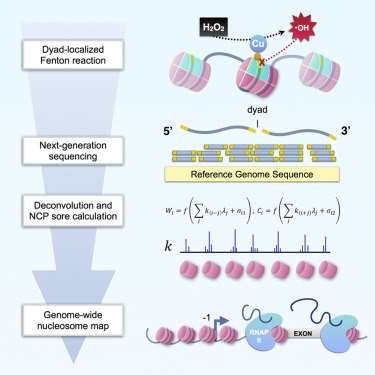
 Our manuscript Insights into Nucleosome Organization in Mouse Embryonic Stem Cells through Chemical Mapping is published in Cell.
Our manuscript Insights into Nucleosome Organization in Mouse Embryonic Stem Cells through Chemical Mapping is published in Cell.
Featured here is our cover illustration entry for the publication.
This cover was fun to make even though we didn’t get selected!
Caption: The nucleosome landscape of a cell can dramatically influence gene expression patterns. In this issue, Voong et al. develop a chemical biology approach to precisely map nucleosomes across the mammalian genome. The cover depicts the chemical mapping technology as a hot air balloon designed in the likeness of a nucleosome. The scientists on board are charting out the nucleosome landscape with a searchlight powered by reagents that catalyze the hydroxyl radical cleavage of nucleosome centers. The authors would like to dedicate the cover art to their former colleague Jon Widom (1955-2011).
 Yanyan’s paper “A Synthetic Biology Approach Identifies the Mammalian UPR RNA Ligase RtcB” is published in Molecular Cell and we are featured on the cover!
Yanyan’s paper “A Synthetic Biology Approach Identifies the Mammalian UPR RNA Ligase RtcB” is published in Molecular Cell and we are featured on the cover!
The unfolded protein response (UPR) maintains homeostasis of the protein-folding environment in the endoplasmic reticulum (ER). Mammalian XBP1 mRNA undergoes unconventional splicing during ER stress. In this issue, using a synthetic genetic circuit, Lu et al. (pp. 758–770) identify RtcB as the long-sought UPR RNA ligase that catalyzes XBP1 splicing in mammals. The illustration takes us to a stressful emergency room where we find RtcB (front surgeon) suturing the spliced XBP1u mRNA (patient) after IRE1 (back surgeon) cleaves and removes intron (sequence shown). The ligation catalyzed by RtcB generates the functional spliced XBP1 (shown in the surgeon’s hands). Read “A Synthetic Biology Approach Identifies the Mammalian UPR RNA Ligase RtcB” here!