Spore-forming bacteria survive on surfaces coated with antimicrobial, latex paints
- Antimicrobial paints kill most bacteria within 24 hours, but some survive
- Such products are typically tested on pathogens but not common bacteria like Bacillus
- Antimicrobial paints could favor antibiotic resistance — making harmless bacteria like Bacillus harmful

SEM image of Bacillus timonensis spores with vegetative cells in the background. Image credit: Jinglin Hu
EVANSTON, Ill. — Antimicrobial paints offer the promise of extra protection against bacteria. But Northwestern University researchers caution that these paints might be doing more harm than good.
In a new study, the researchers tested bacteria commonly found inside homes on samples of drywall coated with antimicrobial, synthetic latex paints. Within 24 hours, all bacteria died except for Bacillus timonensis, a spore-forming bacterium. Most bacilli are commonly found in soil, and many are also found in indoor environments.
“If you attack bacteria with antimicrobial chemicals, then they will mount a defense,” said Northwestern’s Erica Hartmann, who led the study. “Bacillus is typically innocuous, but by attacking it, you might prompt it to develop more antibiotic resistance.”
Bacteria thrive in warm, moist environments, so most die on indoor surfaces, which are dry and cold, anyway. This makes Hartmann question the need to use antimicrobial paints, which may only be causing bacteria to become stronger.
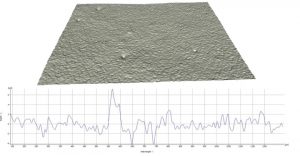
A painted surface that we perceive as smooth is actually quite mountainous on the micro-scale. Image credit: Daniela Ruiz
Spore-forming bacteria, such as Bacillus, protect themselves by falling dormant for a period of time. While dormant, they are highly resistant to even the harshest conditions. After those conditions improve, they reactivate.
“When it’s in spore form, you can hit it with everything you’ve got, and it’s still going to survive,” said Hartmann, assistant professor of civil and environmental engineering in Northwestern’s McCormick School of Engineering. “We should be judicious in our use of antimicrobial products to make sure that we’re not exposing the more harmless bacteria to something that could make them harmful.”
The study was published online on April 13 in the journal Indoor Air.
One problem with antimicrobial products — such as these paints — is that they are not tested against more common bacteria. Manufacturers test how well more pathogenic bacteria, such as E. coli or Staphylococcus, survive but largely ignore the bacteria that people would more plausibly encounter.
“E. coli is like the ‘lab rat’ of the microbial world,” Hartmann said. “It is way less abundant in the environment than people think. We wanted to see how the authentic indoor bacteria would respond to antimicrobial surfaces because they don’t behave the same way as E. coli.”
The study, “Impacts of indoor surface finishes on bacterial viability,” was supported by the Alfred P. Sloan Foundation (award number G-2016-7291) and the Searle Leadership Fund.
This story was originally published by Northwestern University.