The subnuclear localization of DNA is highly regulated in all eukaryotes and has important but poorly understood effects on transcription and chromatin structure. We seek to understand this process by defining the molecular mechanisms that control the positioning of individual genes within the nucleus. In particular, we focus on how yeast genes move from the nucleoplasm to the nuclear periphery to interact with the nuclear pore complex and how this impacts their expression and the spatial organization of the genome as a whole. The mechanisms we have defined in yeast are largely conserved in higher eukaryotes, including humans.
Project 1: How Transcription Factors Control Gene Positioning in Yeast
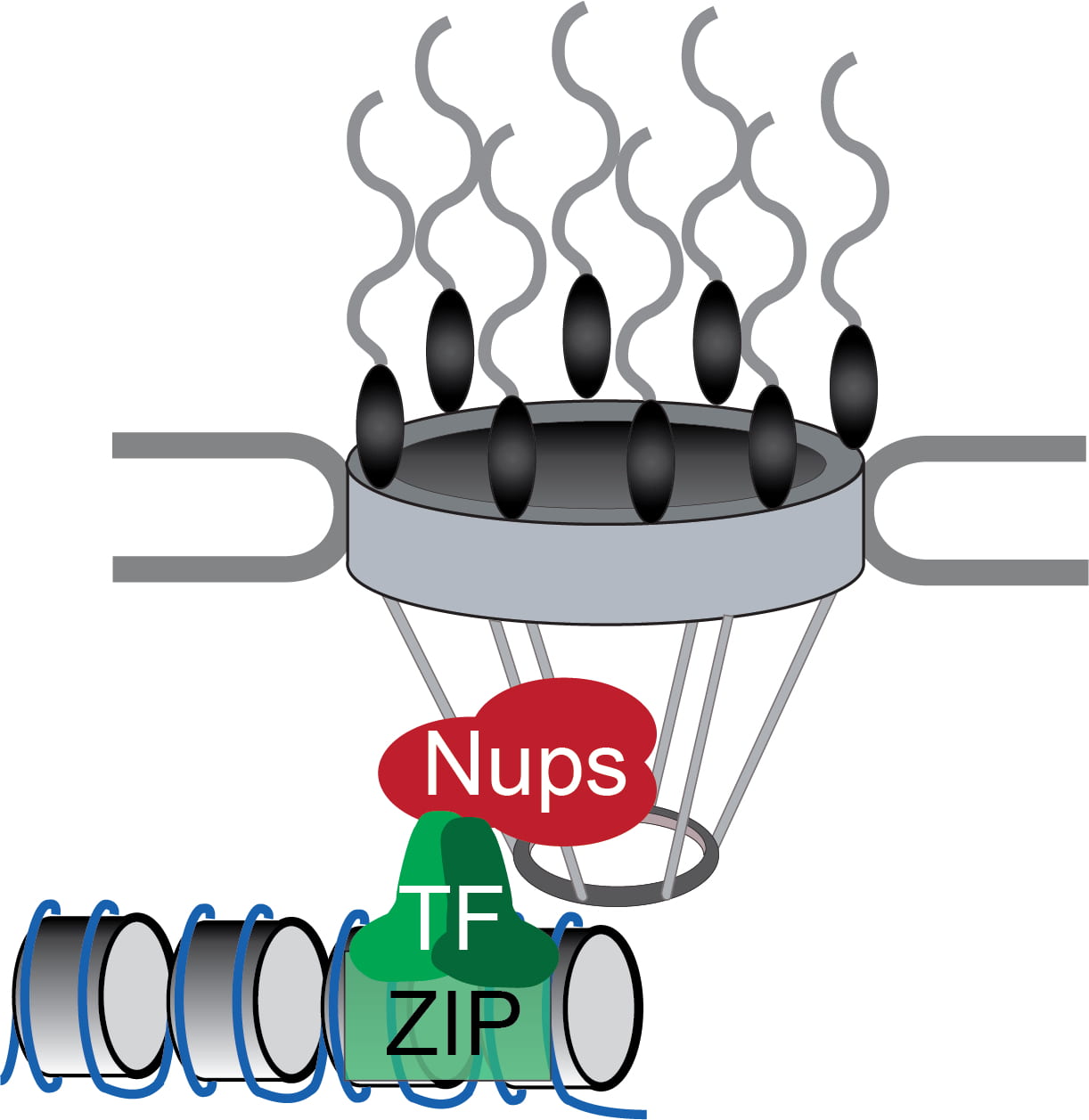
We find that the targeting of genes to the nuclear periphery through interaction with the nuclear pore complex is mediated by small, cis-acting “DNA zip codes” in their promoters. Such zip codes are both necessary for targeting to the periphery and, when inserted at an ectopic site in the genome, sufficient to induce repositioning to the nuclear periphery. These elements are transcription factor binding sites and in many cases we have identified the transcription factors that are responsible for targeting to the periphery. Some, but not all, transcription factors can affect gene positioning. Current work is defining the molecular mechanisms by which transcription factors mediate targeting to the nuclear periphery and comprehensively determining the scope of transcription factor-mediated targeting in yeast.
Project 2: How Gene Positioning Leads to Interchromosomal Clustering
Genes that share DNA zip codes often cluster together within the nucleus and this requires targeting to the nuclear periphery. However, targeting to the nuclear periphery and interchromosomal clustering are separable phenomena that require distinct factors. Current work is focused on determining the global effects of interchromosomal clustering on the genome and determining the molecular mechanism of such interchromosomal interactions.

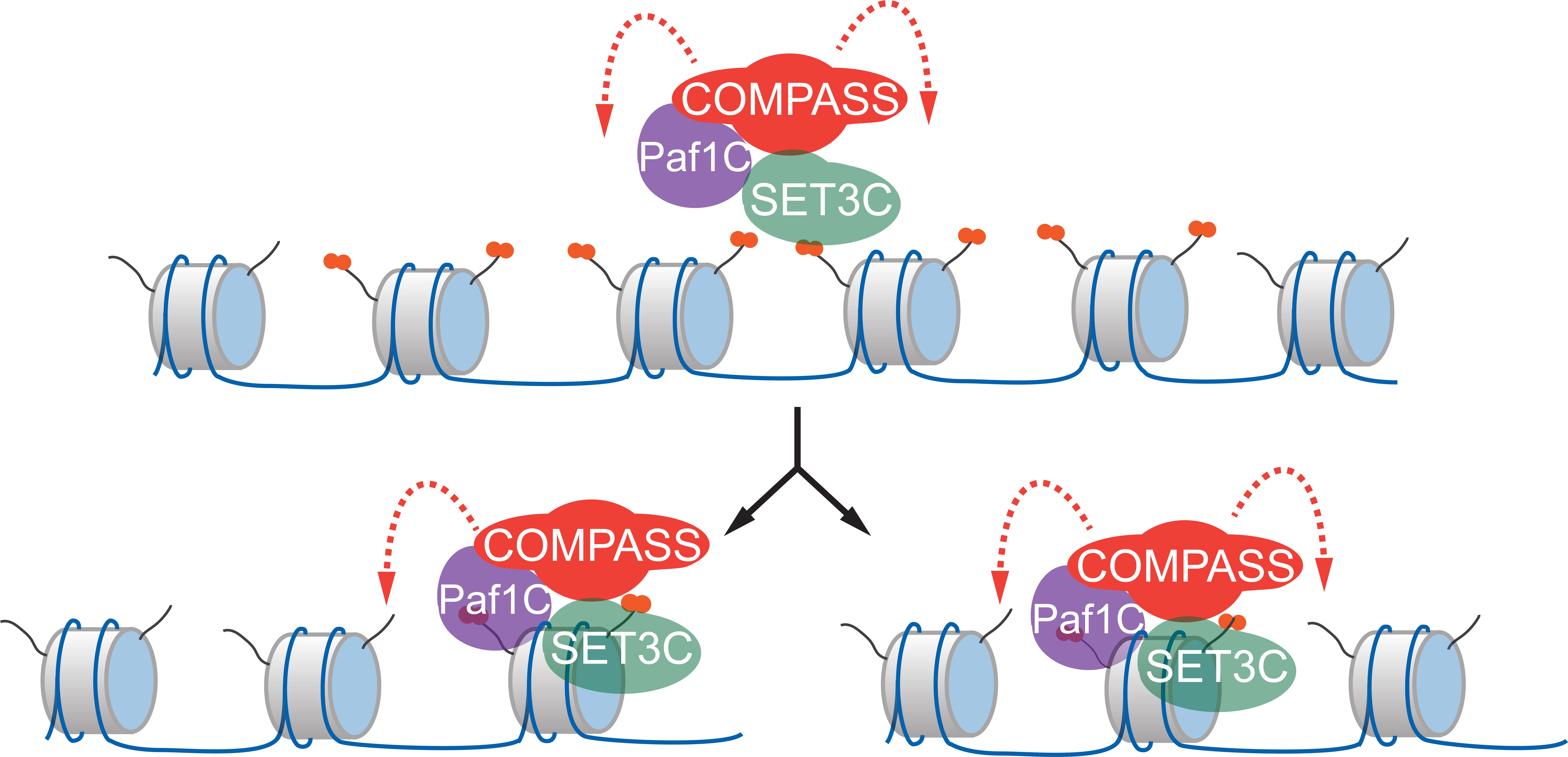
Project 3: How Interaction with the Nuclear Pore Complex Impacts Transcription, Chromatin Structure and Epigenetic Regulatory Mechanisms
We have discovered that at least two different mechanisms target genes to the nuclear pore complex and these mechanisms involve different transcription factors, different nuclear pore proteins and result in different outputs. Most genes that are targeted to the nuclear periphery upon activation rapidly return to the nucleoplasm if they are shut off. However, some genes remain at the nuclear periphery and this localization pattern is maintained through several cell divisions. This second phase, which we call epigenetic transcriptional memory, involves different transcription factors, zip codes and nuclear pore proteins.
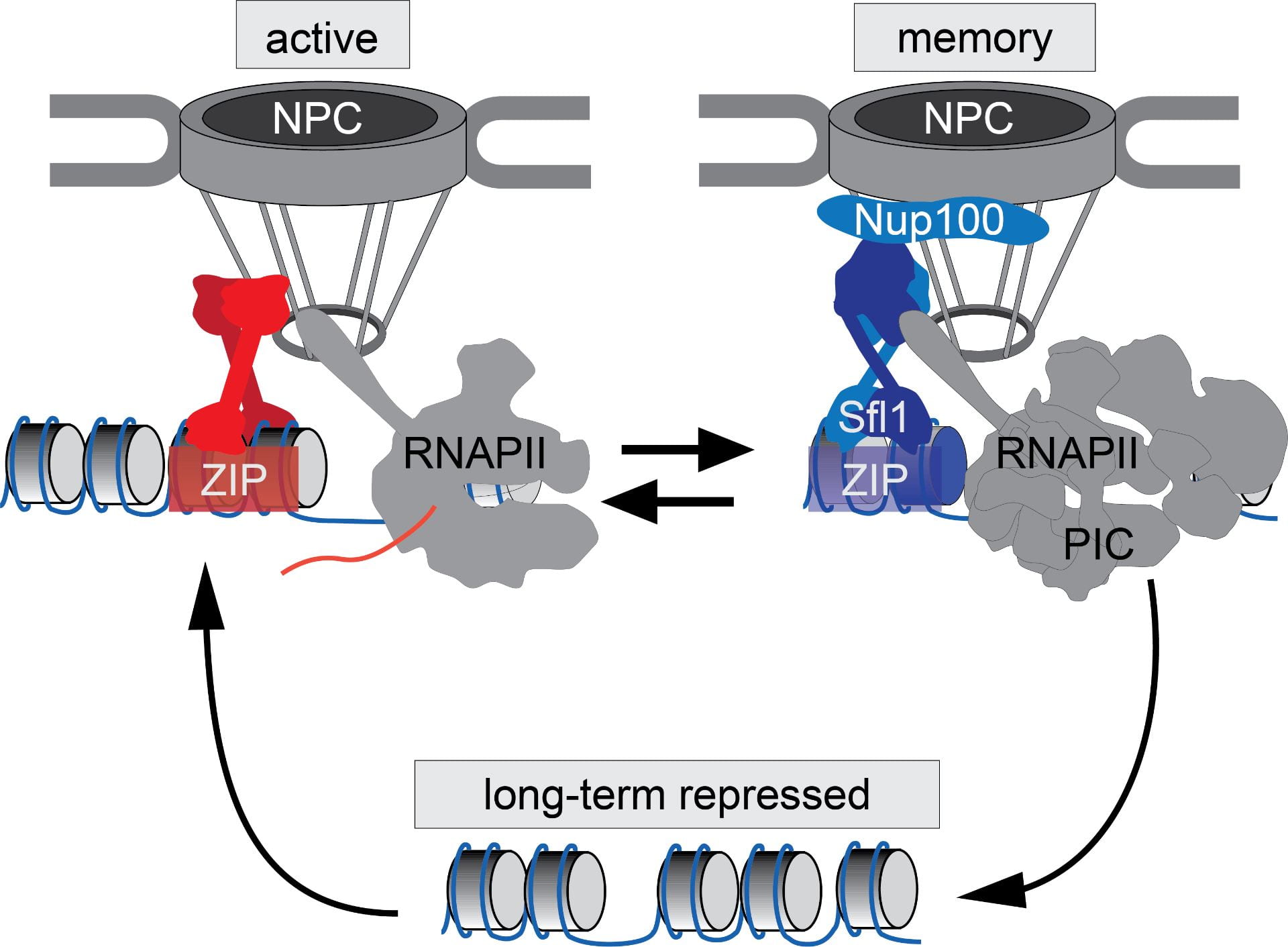
The output of the interaction of active genes with the nuclear pore is generally stronger expression; when the zip codes are mutated or the transcription factors are inactivated, transcription decreases. The output of transcriptional memory is to alter the chromatin structure of the gene, leading to a poised state and increasing the rate of activation in the future. Many yeast and human genes are regulated by a similar mechanism. Currently, we are exploring the physiological roles of transcriptional memory as well as the molecular mechanisms by which the nuclear pore complex impacts both active transcription and transcriptional memory.