Current Collaborations
Physical simulation model of the nucleus – I work closely with physicist Dr. Edward J. Banigan (Prof. Leonid Mirny, MIT) to develop a physical simulation model of the nucleus to investigate the underlying physics of nuclear mechanics and morphology. This physical simulation consists of the two main mechanical components of the nucleus modeled as a polymeric network shell (lamins) and interior polymer (chromatin). Modeling also provides an intriguing avenue to predict biological outcomes before experiments are performed, aiding the pursuit of experiments that will provide significant advances in our understanding. This minimal physical model predicted the nuclear lamina would buckle during extension when the interior chromatin was digested. Upon investigatation by using my micromanipulation technique to extend GFP-lamin A nuclei treated with MNase, buckling was observed experimentally as predicted theoretically. This work was published in my first author paper on nuclear mechanics in Figure 5 and recently published in Biophysical Journal, Mechanics and Buckling of Biopolymeric Shells and Cell Nuclei. Currently we are working on developing this model to investigate the mechanics underlying nuclear bleb formation.
(Stephens et al., 2017 MBoC) (Banigan et al., 2017 Biophys J)
Imaging chromatin density in the nucleus – I have been working with the Prof. Vadim Backman’s lab (Department of Biomedical Engineering at Northwestern) to aid development and application of their novel Partial Wave Spectroscopy (PWS) imaging technique. This label-free imaging technique measures the relative organization of substructures from 20 to 200 nm in size. Together we worked on applying this technique over time to provide temporal changes in nuclear organization as well as the fluctuations of the organization on the scale of 10s of milliseconds. This work is currently in preparation (Gladstein et al.). [ Aim 2B ] – Image below shows formation of a nuclear bleb (arrow) and folding and dynamics of chromatin measured by PWS.
Interrogating Hi-C chromatin interactions and nuclear force response upon digestion of the genome – I am currently collaborating with Prof. Job Dekker (UMass Medical Center) and graduate student Houda Belaghzal to understand how Hi-C chromatin interactions influence nuclear mechanics and how both are altered by differing levels of chromatin digestion. This work is currently being prepared for publication (Belaghzal et al.). [ Aim 1A ]
Light sheet microscopy imaging with concurrent force measurement – I am currently collaborating with Prof. Richard Superfine, Prof. Michael Falvo, and Prof. Timothy O’Brien III and graduate student Chad Hobson (Univeristy of North Carolina – Chapel Hill Departments of Applied Physical Sciences; Physics and Astronomy) on implementing imaging of chromatin during deformation of the cell nucleus due to nuclear force measurement techniques. This collaboration has just begun but is quickly providing novel insights through combining advanced imaging techniques with force measurements. [ Aim 1B ]
(Beicker et al., 2018 Sci Reports)
Screen for chromatin modifying proteins that influence nuclear morphology in a model of breast cancer – I am currently collaborating with Prof. Tanmay Lele and graduate student Andrew Tamashunas (University of Florida – Gainesville Department of Chemical Engineering) on a genetic screen of chromatin proteins that alter nuclear morphology. Abnormal nuclear morphology is common across the human disease spectrum and is most prominently used in cancer for diagnostics. However, we still do not know why these cancers display abnormal nuclear shape and how this alters major functions of the cell nucleus. Initial data from the screen reveals chromatin histone modification enzymes play a substantial role in determining nuclear shape maintenance which is consistent with my recently published paper and front cover art Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. [ Aim 2A ]
Past
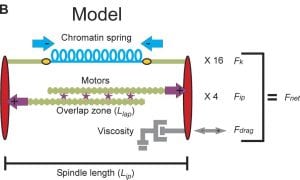
Mathematical model of yeast mitosis – For four years during my graduate work at UNC I led an interdisciplinary collaboration to build a simulation of the yeast mitotic spindle with physicists ( Prof. Michael Falvo), engineers (Prof. Leandra Vicci), computer scientists (Prof. Russ Taylor II and Dr. Cory Quamen) and mathematicians (Prof. Paula Vasquez and Prof. M. Gregory Forest).
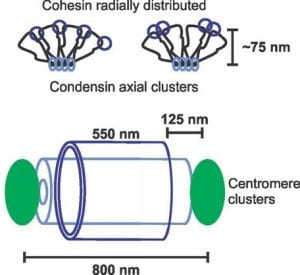
Simulations of fluorescence to determine the fine structure of the yeast pericentromere – During this same period of time I worked hand-in-hand with computer scientists to aid development and provide application of a software that simulates fluorescence of a structure using the experimentally provided point-spread function of the microscope. This collaboration taught me about the physics of light and troubleshooting software under development. In the yeast spindle cohesin (Smc3-GFP) was observed as a barrel structure. This structure provided challenges to understand the fluorescence we were observing. Through building barrels of different sizes and filling them deferentially with fluorophores in simulations provided fine tuned measurements of the size and distribution of structures. This interdisciplinary approach provided a detailed picture of the structure of cohesin, condensin, and pericentromere chromatin that composed the pericentromere chromatin spring in mitosis.